Data
Load the package.
library(CoV19)The data are in the following objects. They have a few common columns: date, region, positive, death. Then there are some other columns depending on the source.
states
italy
worldHere is an example from states:
head(states)## date region positive negative hospitalized death total.tests
## 15132 2020-03-06 AK NA 8 NA 0 8
## 15081 2020-03-07 AK NA 12 NA 0 12
## 15030 2020-03-08 AK NA 14 NA 0 14
## 14979 2020-03-09 AK NA 23 1 0 23
## 14928 2020-03-10 AK NA 23 1 0 23
## 14877 2020-03-11 AK NA 46 1 0 46Subsetting data
# Just WA
x <- subset(states, region=="WA")
# Two states
x <- subset(states, region %in% c("WA","CA"))
# All areas in China
x <- subset(world, stringr::str_detect(region, "China"))Merging Data
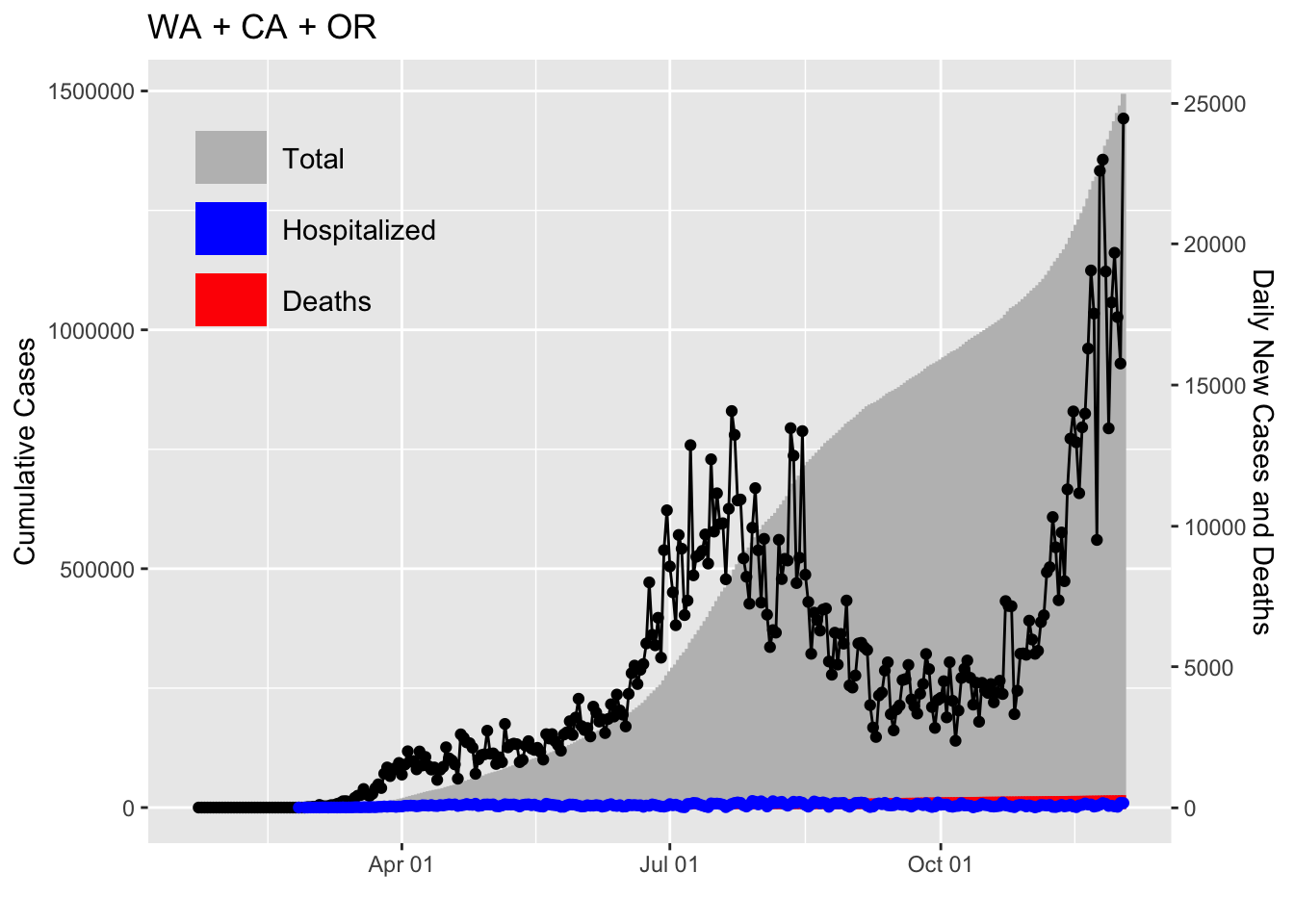
Let’s say you want to have the sums for all regions. You can do that with dplyr.
library(dplyr)
# If you are unfamiliar with dplyr, the %>% is a pipe that sends
# the result to the left into the function in the right
x <- states %>%
subset(region%in%c("WA","CA","OR")) %>%
group_by(date) %>%
summarize_if(is.numeric, sum, na.rm=TRUE)
# This will not have the region column, so we add that back on
x$region <- as.factor("WA + CA + OR")We can pass this data object to plot2 to plot.
plot2(x)